Corrections
Below is a running list of minor corrections for our papers (paper numbers correspond to previous page). If you identify any other issues, please send us an email!
29.
Dollo-CDP: a polynomial-time algorithm for the clade-constrained large Dollo parsimony problem; available at doi: 10.1186/s13015-023-00249-9
Use of \(V(T)\)
Sometimes \(V(T)\) denotes the set of internal (i.e., non-leaf) vertices in a phylogenetic tree \(T\) and sometimes it denotes all vertices in \(T\). The usage is clear from the context.
Definition of \( \mathcal{STB} \)
On page 7, the definition of \( \mathcal{STB} \) should be \( \mathcal{STB} = \{ X|Y : X,Y,X \cup Y \in \Sigma \text{ and } X \cap Y = \emptyset \} \). The definition in the paper is missing \( X \cap Y = \emptyset \); however, this condition is clear from the context.
Algorithm 3 in Additional file 1
The algorithm description is inconsistent with the main text and the Dollo-CDP code. Based on the main text, there should be another conditional after line 7 to check if \(Y = A \setminus X\) is a member of \(\Sigma\) before appending \(X\) to \(SubBip[A]\). If using bitvectors, membership in \(\Sigma\) can be checked with an \(O(n)\) hash. This is the same as checking if \(X\) is a proper subset of \(A\) so it does change our time complexity analysis.
27.
Quartets enable statistically consistent estimation of cell lineage trees under an unbiased error and missingness model; available at doi: 10.1186/s13015-023-00248-w
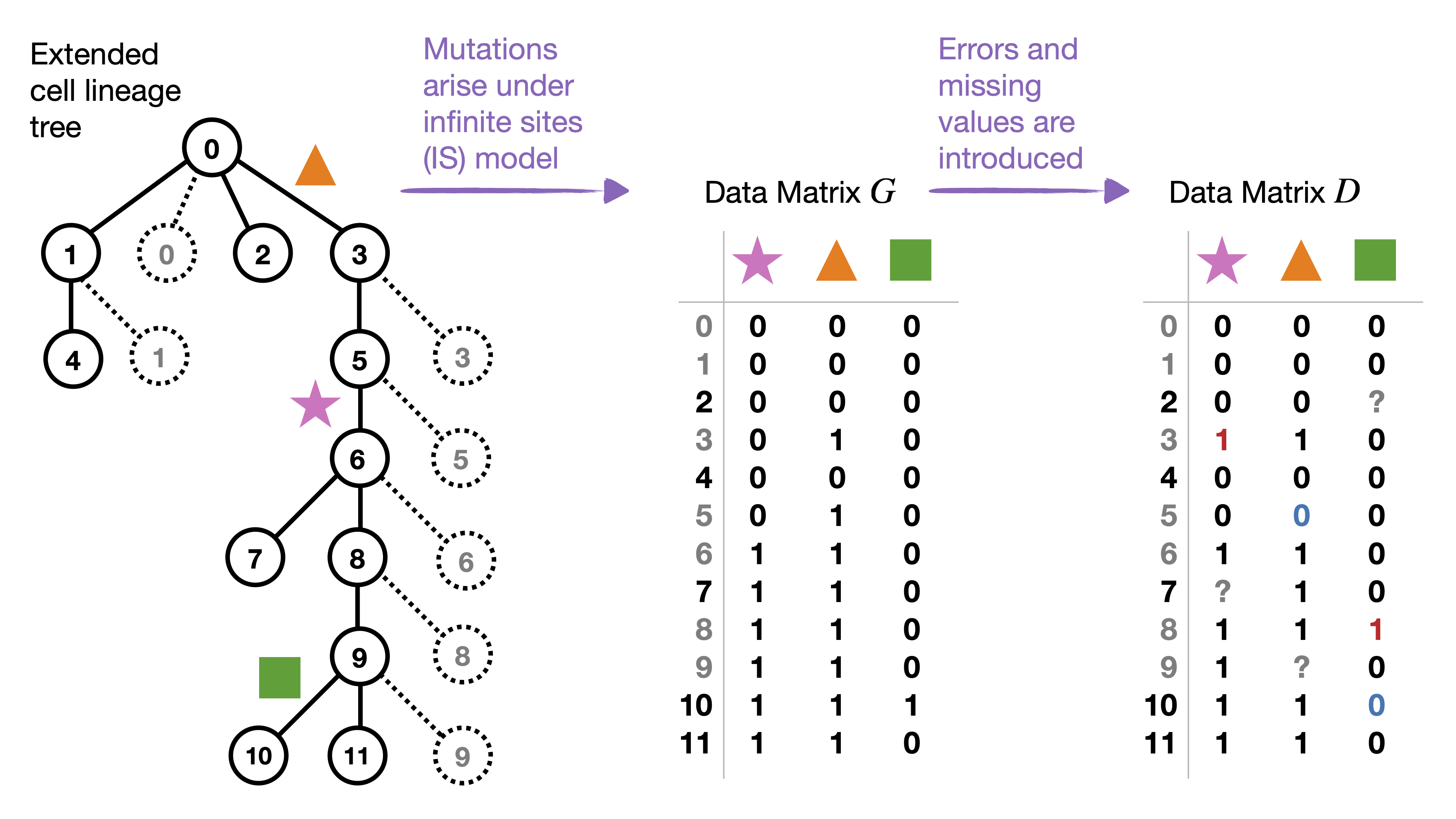
Figure 1
The entry of \(G\) corresponding to cell 4 (row) and orange triangle mutation (column) should be set to 0 instead of 1 because cell 4 is not below the orange triangle mutation in the cell lineage tree so it does not inherit the mutation from an ancestor (see corrected figure below). This typo does not impact any other part of the paper.
23.
Theoretical and practical considerations when using retroelement insertions to estimate species trees in the anomaly zone; available at doi: 10.1093/sysbio/syab086
Definition of false positive rate (page 726)
False positive rate should be defined the number of branches in the estimated species tree that are missing from the true tree, divided by the number of internal branches in the estimated tree (instead of true tree).
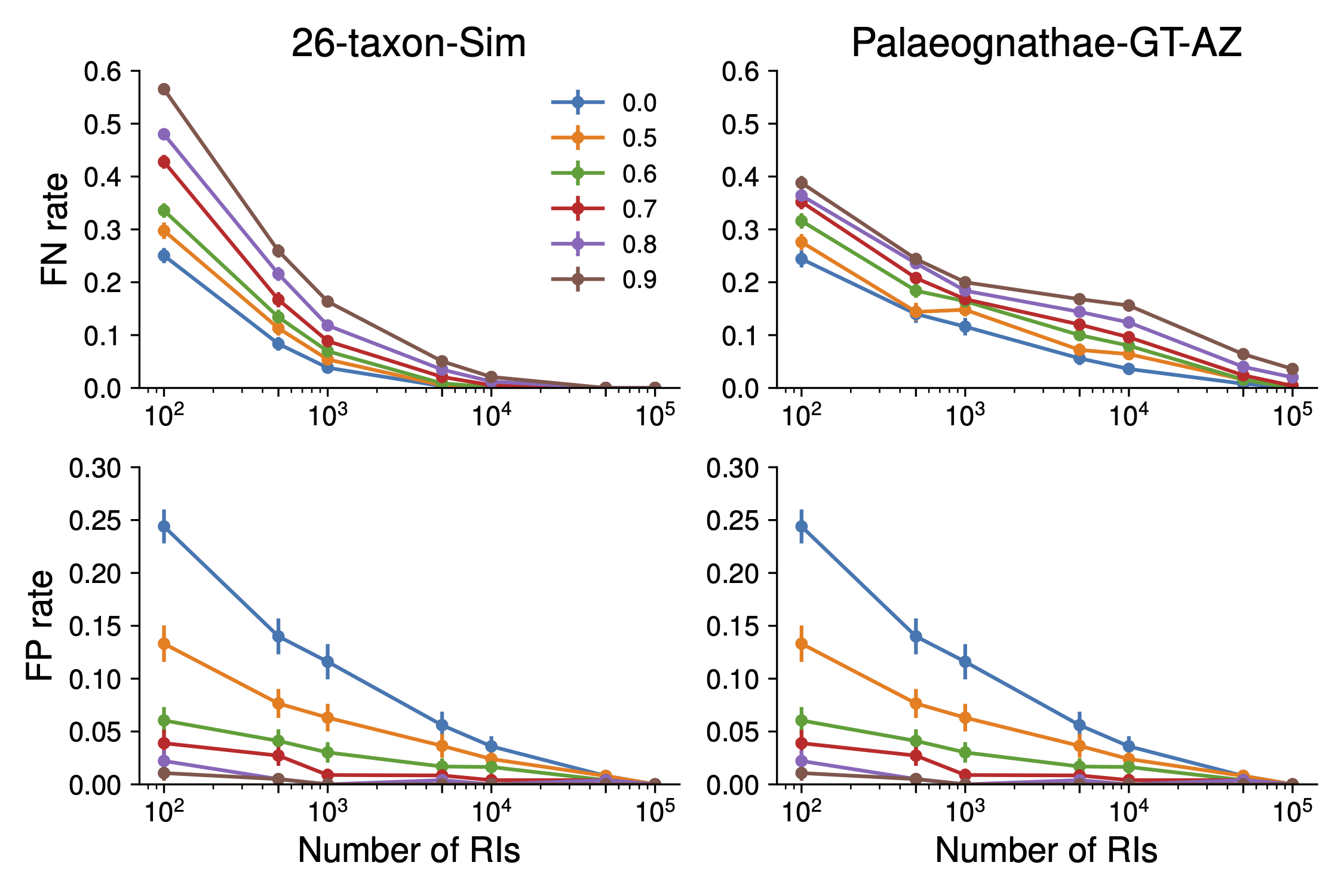
Supplementary Figure S1
The mistake above was made in both the text and our analyses. We corrected our analyses on Github. There are no noticeable differences between the corrected figure below and Supplementary Figure S1; therefore, this error does not change our results or conclusions.
15.
FastMulRFS: fast and accurate species tree estimation under generic gene duplication and loss models;
available at doi: 10.1093/bioinformatics/btaa444
Equation (2)
The second \(+\) should be \(-2\) so that the equation reads
$$
RF(T, T') = |E(T|_R)| + |E(T')| - 2 | C(T|_R) \cap C(T') |
$$
where \(T\) is a phylogenetic tree on label set \(S\), \(T'\) is a tree on label set \(R \subseteq S\), \(E(T)\) returns the edges of \(T\), and \(C(T)\) returns the bipartitions of \(T\).
The correct equation is used in the proof on page i64, so this typo does not impact our results.
9.
To Include or Not to Include: The Impact of Gene Filtering on Species Tree Estimation Methods; available at doi: 10.1093/sysbio/syx077
Broken reference (page 287)
The broken reference \(?\) should point to Kubatko and Degnan, Systematic Biology, 2007 doi: 10.1080/10635150601146041.